Wing-Cheung YAM, Kenny CW CHAN, SS LEE
Treatment of HIV infection has been revolutionalised by the regular use of highly active antiretroviral therapy (HAART). This is supported by the measurement of CD4 count and viral loads, which are now the standard monitoring tools in clinical services where they are accessible. These routine investigations serve to track progress and inform responses. To further optimise current therapy, new investigative tools are introduced, some of which are fast becoming new standards. Resistance testing is an investigation that has evolved from an experimental tool to a clinical measure, the goal of which is to predict treatment response. This is paralleled by therapeutic drug monitoring (TDM), a pharmacological investigation the principles of which are not too different from the determination of antibiotic levels in the monitoring of treatment of bacterial infections. This chapter is devoted to a description of these new tools, and is concluded with a discussion of the application of viral fitness testing in future. An algorithm at the end of the chapter gives a overview of the potential roles of these new investigations.
There are two main forms of HIV resistance: Primary or transmitted resistance refers to that arising from the infection by a virus that is already resistant to selected antiretrovirals. The prevalence of primary resistance varies from one population to another. The other form is secondary or acquired resistance, a condition that evolves from exposure to antiretroviral drugs.
Acquired resistance has gradually become an important clinical observation. In the past decade, highly active antiretroviral therapy (HAART) has lowered morbidity and mortality of HIV infection. A proportion of patients on HAART does not achieve optimal viral suppression or may experience viral rebound within a short period. HAART induced mutations in HIV-1 reverse transcriptase (RT) and protease (PT) allows viral escape from drug suppression, a phenomenon that is becoming well characterised. On the other hand, the determination of primary resistance is driven by public health needs through the establishment of surveillance mechanisms. Over the years, nucleoside reverse transcriptase inhibitor (NRTI) resistance has fluctuated over time and across geographic areas in western countries, from 10% to over 20%. Studies suggested that PI resistance is stabilising whereas non-nucleoside reverse transcriptase inhibitor (NNRTI) resistance has been on the rise, to over 10% in recent years.1,2 The prevalence of primary resistance to 2 or more classes of antiretroviral drugs has remained low at <5%.1
The nature, extent and potential impacts of resistance-associated failure cannot be inferred from conventional routine laboratory tests (CD4 and viral load). Studies in United States and Europe have found that the application of HIV resistance testing could improve treatment response to HAART. This has led international authorities to recommend on the incorporation of resistance testing into patient management in some settings.3
Antiretroviral resistance can be detected using either genotypic or phenotypic assays. HIV-1 genotypic assays identify mutations or changes in the nucleotide sequences that are known to confer decreased susceptibility to antiretroviral drugs. The phenotype is a trait or behaviour resulting from the expression of a specific genotype. HIV-1 phenotypic assays measure the ability of virus to replicate in the presence of antiretroviral drugs. Results of phenotypic assays are typically reported as the inhibitory concentration (IC50), which is the concentration of the tested drug that reduces in vitro HIV-1 replication by 50%. These assays involve highly complex protocols and analytical procedures undertaken in sophisticated laboratory environment. Todate, only a small number of specialised clinical reference laboratories around the world have the facilities and technical expertise to provide reliable phenotypic drug susceptibility results in the clinically relevant time frame. Phenotypic testing is currently not available in Hong Kong as a regular service in clinical HIV management.
HIV-1 genotypic assay involves the determination of nucleotide sequences of the reverse transcriptase and protease genes of the virus. Sequence changes that are known to confer resistance to antiviral drugs are catalogued. Point mutations associated with drug resistance may affect the fitness or ability of the virus to replicate its genome. Resistance mutation is often classified as major or minor forms. Major resistance mutations impact drug susceptibility and allow the virus to survive and replicate in the presence of antiretroviral. Minor resistance mutations are less clinically relevant. They normally appear after major mutations to compensate for changes that have been generated. Genotypic analysis of plasma HIV RNA involves several steps: extraction and purification of total RNA, reverse transcription, amplification of HIV DNA sequences encompassing the RT and PT regions, and sequencing of amplified products for detection of mutations associated with antiretroviral resistance. In Hong Kong genotyping resistance testing was introduced as a pilot in 2001.4
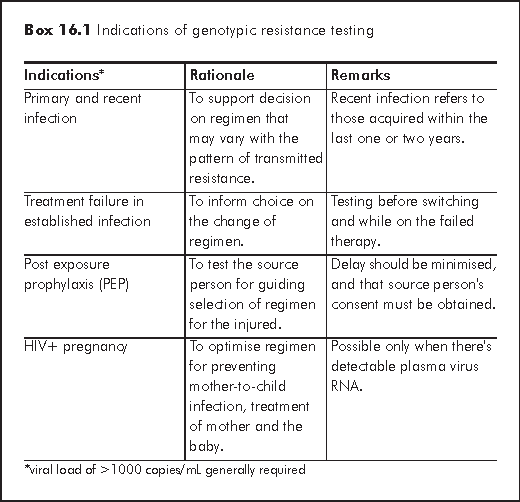
Compared to phenotyping assays, genotyping resistance testing is more commonly used in support of clinical HIV management. Clinical guidelines have been established by various authorities, including the International AIDS Society-USA Panel,3 Euroguidelines Group for HIV Resistance5 and DHHS Panel on Antitretroviral Guidelines for Adults and Adolescents.6 A summary of the general indications for genotypic resistance testing is at Box 16.1. Recent study has shown that use of resistance testing to determine the next regimen following treatment failure is cost-effective.

In addition, the use of resistance testing to guide the initial treatment regimen appears to be cost-effective if the prevalence of primary genotypic resistance in the population is at least 4%.7
Currently, four general classes of antiretroviral drugs are used in clinical care, nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), protease inhibitors (PIs), and entry inhibitors. Viral resistance can occur with each of these classes of drug, particularly when viral replication is not maximally suppressed while on therapy. This may arise from non-adherence to HAART, and suboptimal treatment like monotherapy or two drug-combinations, which are no longer used in clinical practice.
Genotypic assays involve two independent processes, identification of resistance mutations and interpretation of how these mutations alter HIV-1 drug susceptibility. An error in either process may lead to an inaccurate genotyping results. Currently two types of interpretation systems have been designed by various authorities. Firstly, the 'rules-based' systems are deduced by experts, from clinical experiences and literature. In addition to listing the mutations identified in the RT and protease genes, an interpretative report is provided that lists each drug and provides a designation of either "susceptible", "low-level resistance", "intermediate resistance", or "high-level resistance". "Susceptible" is used if no known mutations are detected or if reduced susceptibility to a specific drug has not been associated with the mutation. "Intermediate resistance" is used when the mutation detected have been associated with diminished virological responses in some but not all patients. "High-level resistance" refers to mutations that have been associated with a maximum reduction in susceptibility to the drug. This rules-based system provides clinicians with results in a user-friendly format that can be easily understood without the need for an extensive knowledge of the genetics of HIV-1 resistance. The Los Alamos Database http://www.hiv.lanl.gov/content/hiv-db/ADRA/adra.html and Stanford database http://hivdb.stanford.edu are two of the examples.3,8,9 On the other hand, there's the "database-driven" system with algorithms derived from bioinformatics approaches. Virtual PhenotypeTM is an interpretation system that predicts phenotypes from genotypes by comparing the query sequence with those available in the database and through averaging the resistance of the matching sequences.
A list of mutations is maintained by the International AIDS Society-USA Drug Resistance Mutations Group, which is brought updated from time to time. A list of common mutations is at Box 16.2. The main features of the resistance patterns of the three main classes of antiretrovirals are:
NRTI - TAM (thymidine analogue mutation) or NAM (nucleoside analogue mutations) include a number of mutations that commonly occur in various NRTIs. Reduction of virological responses is proportional to the number of TAMs or NAMs. Cross resistance is the rule. The lamivudine/emtricitabine specific resistance M184V is, however, a single mutation that renders a high degree of resistance. Reduction of virus replication may occur with some mutations like M184V, K65R. Some mutations like K65R can increase the sensitivity to another antiretroviral (zidovudine).
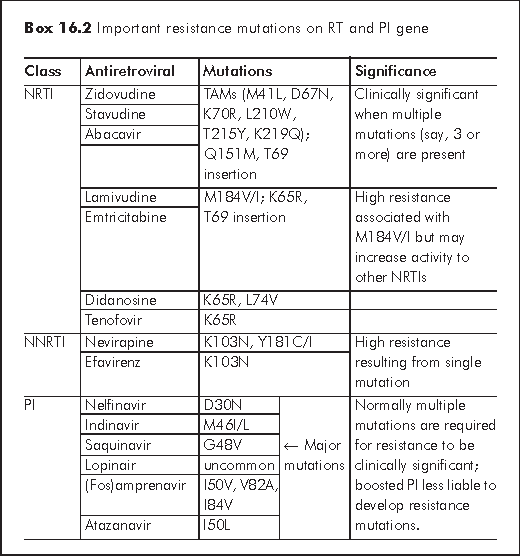
NNRTI - The common characteristics of NNRTI is the propensity for high resistance after a single mutation. K103N is the common mutation which, if present, leads to cross resistance to other NNRTIs. Interestingly, NRTI experienced patients could present with hypersusceptibility to NNRTI.
PI - Primary mutations are relatively inhibitor specific and may alter the viral susceptibility to the drug. Primary resistance mutations associated with the use of boosted PI is generally less common than unboosted PI. Clinically significant resistance occurs when there's accumulation of multiple mutations. Secondary or minor mutations alone may have no effect on viral susceptibility but may improve viral fitness, allowing a virus with a primary mutation to improve its replicative capacity. Polymorphisms at some of the positions may in fact compensate for the reduced protease activity. M36I, for example, is a natural polymorphism that is common in non-B HIV, an observation that has also been confirmed in Hong Kong.4
Other than stavudine and didanosine, antiretrovirals are typically prescribed for adults in standard dosage regardless of patient characteristics such as body weight and gender. Certain standard dose adjustments are recommended for anticipated drug-drug interactions, such as between protease inhibitors and rifamycins. However, patient characteristics are still not considered.
It is now known that considerable inter- and intra-individual antiretroviral levels occur, even without concurrent interacting drugs. Furthermore, for some drugs, plasma concentrations have been associated with occurrence of adverse effects, and levels achieved with standard dosage for some drugs are only marginally higher than the required inhibitory concentrations. Such inhibitory concentrations themselves also vary significantly with the accumulation of resistant mutations.
Based on these observations, some overseas centres have employed therapeutic drug monitoring (TDM) to assist therapeutic decisions.10 In Hong Kong, it is available only on a research basis. TDM is typically performed on plasma by high performance liquid chromatography coupled with spectrofluorometric detection. As with some antibiotics, Cmin and Cmax are used to monitor treatment efficacy and drug toxicity. However, for some drugs, the Area Under Curve (AUC) may be more predictive. With resistant virus, the relationship between the plasma level and IC50 (the inhibitory quotient, IQ) is used to predict efficacy of non-standard regimens or determine dosage of conventional regimens.11 The current success of boosted-PI and NNRTI-based regimens have been attributed to a favourable IQ, among other factors. Some studies have in fact successfully correlated drug exposure with short-term treatment success in patients with resistant virus.12
TDM is not yet recommended for routine clinical use. Currently, among the three main classes of antiretrovirals used, only PI and NNRTI can be assayed for their plasma levels. NRTI, though an important backbone of standard HAART, is activated intracellularly. Its plasma concentration is not meaningful and very little data are available with intracellular monitoring of NRTI levels as a clinical tool. Technically, it is cumbersome and time-consuming to perform a full pharmacokinetic profile. Consensus is not yet achieved on a protocol of simplified sampling method for all drugs. A lag time of at least 2 weeks is required for steady state concentrations to be achieved; by then, it may be too late to reverse some clinical decisions. TDM also suffers from the following limitations in clinical applications:
(a) The body of knowledge regarding the relationship between treatment outcome and various pharmacokinetic parameters such as Cmax, Cmin, Tmax (time to peak concentration) and AUC is still incomplete. Attempts are currently being made to establish the therapeutic range and IQ.
(b) Current antiretroviral drugs are only available in limited denominations, precluding fine adjustment of dosage.
(c) There are limited supportive data on the long term clinical benefit of adopting TDM, especially in treatment-experienced patients.13,14
Despite these concerns, an increasing number of HIV treatment centres have adopted testing, especially in Europe. Currently, TDM may be considered in the following situations:
(a) Suspected non-adherence - TDM showing very low drug levels may provide supportive evidence.15
(b) Unpredictable drug-drug interactions - such as the combined use of amprenavir and boosted lopinavir.16
(c) Unpredictable pharmacokinetics - such as in pregnant women or patients with liver failure, when dose adjustment may be advantageous
(d) Management of side effects - for supporting the diagnosis and possible titration of dosages.
Replicative capacity (RC) is the laboratory measurement of viral fitness, a term that describes the adaptability of HIV in a given environment, typically in competition with other strains. Viral fitness may differ according to subtypes, accounting for the differential spread in different parts of the world.17 It may also differ according to eras, leading some to suggest that HIV has attenuated over time.18
At the clinical level, viral fitness is important as it may decrease as mutations develop in response to the selection pressure exerted by antiretrovirals. As the selection pressure continues, secondary mutations usually develop to restore viral fitness and presumably renewed pathogenicity. However, until then, a discord between a detectable viral load and a stable CD4 count occurs.
In most cases, it is impossible to predict how long this discord will persist before restored viral fitness can lead to a fall in CD4 count. Yet in patients with a borderline CD4 count and limited treatment options, the ability to predict and hence switch therapy to maintain this viral load-CD4 discord is theoretically useful.
There is some evidence that baseline RC predicts subsequent disease progression.19 In treatment-experienced patients on salvage therapy, RC also correlates with CD4 count and viral load.20 However, in a setting with access to resistance testing and viral load, it is unclear how RC will affect the clinical outcome. It is also premature to introduce RC into clinical practice, there being no standard assay of viral fitness. Although the principle remains one of comparing the fitness against that of wild type, different methodologies have been used. In one, plasma-derived RT and PR sequences are inserted into a retrovirus vector containing a luciferase indicator gene. RC is then determined by measuring luciferase activity in infected cells after a single round of replication, as % of wild type strain. In another, the virus is simply co-cultured with wild type virus.
Currently, the use of RC in Hong Kong is limited to research settings and cannot be recommended for routine clinical use.
HIV-1 non-B viruses, especially CRF01_AE has become the predominant HIV-1 subtype circulating in Hong Kong and in some other Asian developing countries. However, most of the published data on the treatment responses to antiretrovirals refer to B subtypes. In addition, many of the virologic diagnostic tests, for example, phenotypic and genotypic HIV-1 drug resistance assays, have been derived largely from studies on subtype B viruses. This has arisen from the predominance of drug therapy in those areas of the world in which subtype B viruses predominate. The applicability and validity of the available knowledgebase to non-B subtypes may need to be examined. The same challenge also exists when newer tests like TDM and replicative capacity gradually become introduced. There is a need for new algorithms and interpretation systems for supporting the optimisation of clinical management of non-B HIV subtype infected populations, including Hong Kong and many countries in the developing world.
Grant RM, Hecht FM, Warmerdam M, et al. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA 2002;288:181-8.
Cane P, Chrystie I, Dunn D, et al. Time trends in primary resistance to HIV drugs in the United Kingdom: multicentre observational study. BMJ 2005;331:1368.
Hirsch MS, Brun-Vezinet F, Clotet B, et al. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA Panel. Clin Infect Dis 2003;37:113-28.
Yam WC, Chen JH, Wong KH, et al. Clinical utility of genotyping resistance test on determining the mutation patterns in HIV-1 CRF01_AE and subtype B patients receiving antiretroviral therapy in Hong Kong. J Clin Virol 2006;35:454-7.
EuroGuidelines Group for HIV Resistance. Clinical and laboratory guidelines for the use of HIV-1 drug resistance testing as part of treatment management: recommendations for the European setting. The EuroGuidelines Group for HIV resistance. AIDS 2001;15:309-20.
DHHS Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1 infected adults and adolescents. October 10, 2006. Available from http://aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf (accessed 12 December 2006).
Weinstein MC, Goldie SJ, Losina E, et al. Use of genotypic resistance testing to guide hiv therapy: clinical impact and cost-effectiveness. Ann Intern Med 2001;134:440-50.
Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1: Fall 2005. Top HIV Med 2005;13:125-31.
Hanna GJ, D'Aquila RT. Clinical use of genotypic and phenotypic drug resistance testing to monitor antiretroviral chemotherapy. Clin Infect Dis 2001;32:774-82.
Haubrich R, Demeter L. International perspectives on antiretroviral resistance. Clinical utility of resistance testing: retrospective and prospective data supporting use and current recommendations. J Acquir Immune Defic Syndr 2001;26 Suppl 1:S51-9.
Winston A, Hales G, Amin J, et al. The normalized inhibitory quotient of boosted protease inhibitors is predictive of viral load response in treatment-experienced HIV-1-infected individuals. AIDS 2005;19:1393-9.
Baxter JD, Merigan TC, Wentworth DN, et al. Both baseline HIV-1 drug resistance and antiretroviral drug levels are associated with short-term virologic responses to salvage therapy. AIDS 2002;16:1131-8.
Bossi P, Peytavin G, Ait-Mohand H, et al. GENOPHAR: a randomized study of plasma drug measurements in association with genotypic resistance testing and expert advice to optimize therapy in patients failing antiretroviral therapy. HIV Med 2004;5:352-9.
Clevenbergh P, Garraffo R, Durant J, Dellamonica P. PharmAdapt: a randomized prospective study to evaluate the benefit of therapeutic monitoring of protease inhibitors: 12 week results. AIDS 2002;16:2311-5.
Hugen PW, Burger DM, Aarnoutse RE, et al. Therapeutic drug monitoring of HIV-protease inhibitors to assess noncompliance. Ther Drug Monit 2002;24:579-87.
Basso S, Solas C, Quinson AM, et al. Pharmacokinetic interaction between lopinavir/r and amprenavir in salvage therapy. J Acquir Immune Defic Syndr 2002;31:115-7.
Arien KK, Abraha A, Quinones-Mateu ME, et al. The Replicative fitness of primary human immunodeficiency virus Type 1 (HIV-1) Group M, HIV-1 Group O, and HIV-2 isolates. J Virol 2005;79:8979-90
Arien KK, Troyer RM, Gali Y, Colebunders RL, Arts EJ, Vanham G. Replicative fitness of historical and recent HIV-1 isolates suggests HIV-1 attenuation over time. AIDS 2005;19:1555-64.
Daar ES, Kesler KL, Wrin T, et al. HIV-1 pol replication capacity predicts disease progression. AIDS 2005;19:871-7.
Sarmati L, Nicastri E, Montano M, et al. Decrease of replicative capacity of HIV isolates after genotypic guided change of therapy. J Med Virol 2004;72:511-6.